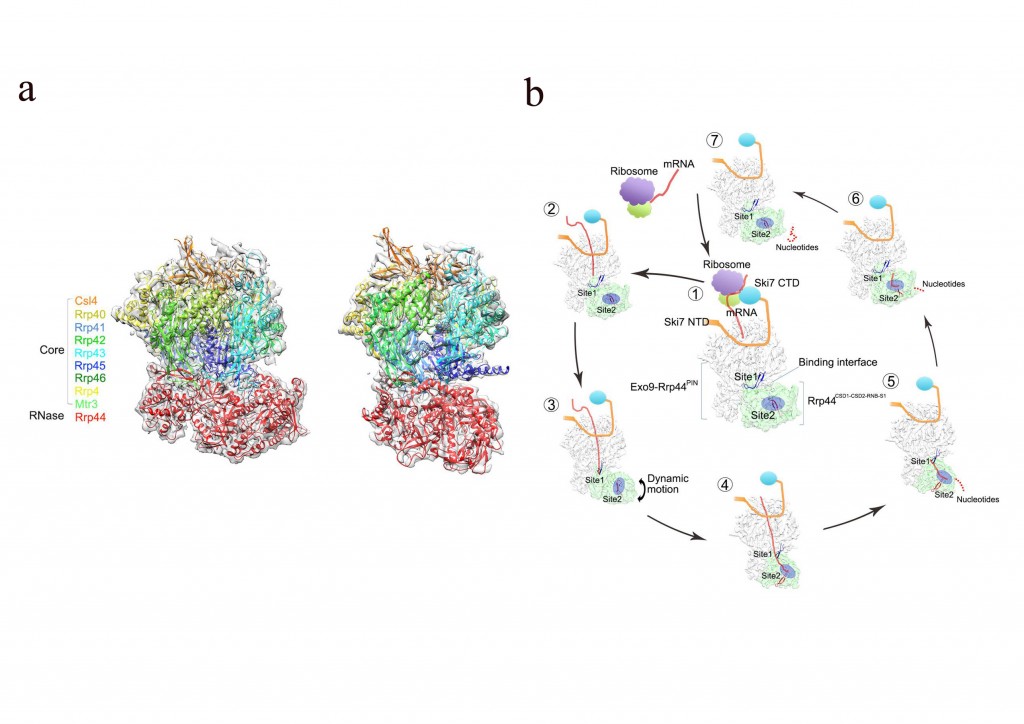
1) The multi-subunit exosome complex is a key player in RNA quality control systems of eukaryotic cells. In order to fully understand the mechanisms and regulations of this important cellular machinery, we utilize cryo-EM and single particle reconstruction techniques to capture clear snapshots of the entire exosome complex binding with its different cofactors in various working states.

Liu JJ, Bratkowski MA, LiuX, Niu CY, Ke A*, and Wang HW*. (2014) Visualization of distinct substrate-recruitment pathways in the yeast exosome by EM. Nat. Struct. Mol. Biol., 21(1):95-102 (*co-corresponding authors)
2) Recently, we used single-particle cryo-electron microscopy to solve the structures of
the Ski7-exosome complex in RNA-free and RNA-bound forms at resolutions of 4.2 Å and 5.8 Å, respectively. These structures revealed that the N-terminal domain of Ski7 adopts a structural arrangement and interacts with the exosome in a similar fashion to the nuclear Rrp6’s C-terminal domain. Further structural analysis of the exosome with different RNAs suggests a switch mechanism of RNA-induced exosome’s activation in through-core RNA processing.